Workflow
How you organise yourself when doing R will make it easier for you if you need to go back to your script weeks or years after, and to share with others people.
There is different workflow that you can do. At first, when I was
working on a project, I was using a simple folder named analyses and
put everything in it with obscure name such as spatial analyses.R or
canopy area analyses.R, etc. I was also putting all the outputs
(Figures, tables, maps) in the same working directory as the .R scripts.
I think, this make it harder to find again some figures that you did made.
Now I am using since a couple of year another workflow. I first used
number before each names to know which script I need to run at first
like 1 data upload.R, 2 function point pattern.R and 3 final
results.R.
For the outputs, I have .RData saving files in the working directory,
but all my Figures are in different folder. Thus, for example, I have
a path like this ./living/NA/normal/replicate/Lmm graph/ with every
figures of the L_mm functions (a spatial pattern function) for the
replicate normal NA (no marks) and living patterns.
I have a little script that help in R for checking if the path is here and make a new folder or not if it already exist.
Here is the little script will check for the path /Project X/Analyses/results RandomForest:
mainDir <- "/Project X/Analyses"#path do the main folder without "/" to the end
if (file.exists(file.path(mainDir, "results RandomForest"))) {
setwd(file.path(mainDir, "results RandomForest"))
} else {
dir.create(file.path(mainDir, "results RandomForest"), showWarnings = FALSE,recursive=TRUE)
setwd(file.path(mainDir, "results RandomForest"))
}
You can even add it in a for loop like this:
mainDir <- "/Project X/Analyses"#path do the main folder without "/" to the end
#you can use here
# mainDir <- getwd() # to have this script work
setwd(mainDir)
variables_names <- c("temperature","slope","patterns")
for (i in variables_names) {
library(ggplot2)
library(ggthemes)
if (file.exists(file.path(mainDir, "results RandomForest",i))) {
setwd(file.path(mainDir, "results RandomForest",i))
} else {
dir.create(file.path(mainDir, "results RandomForest",i), showWarnings = FALSE,recursive=TRUE)
setwd(file.path(mainDir, "results RandomForest",i))
}
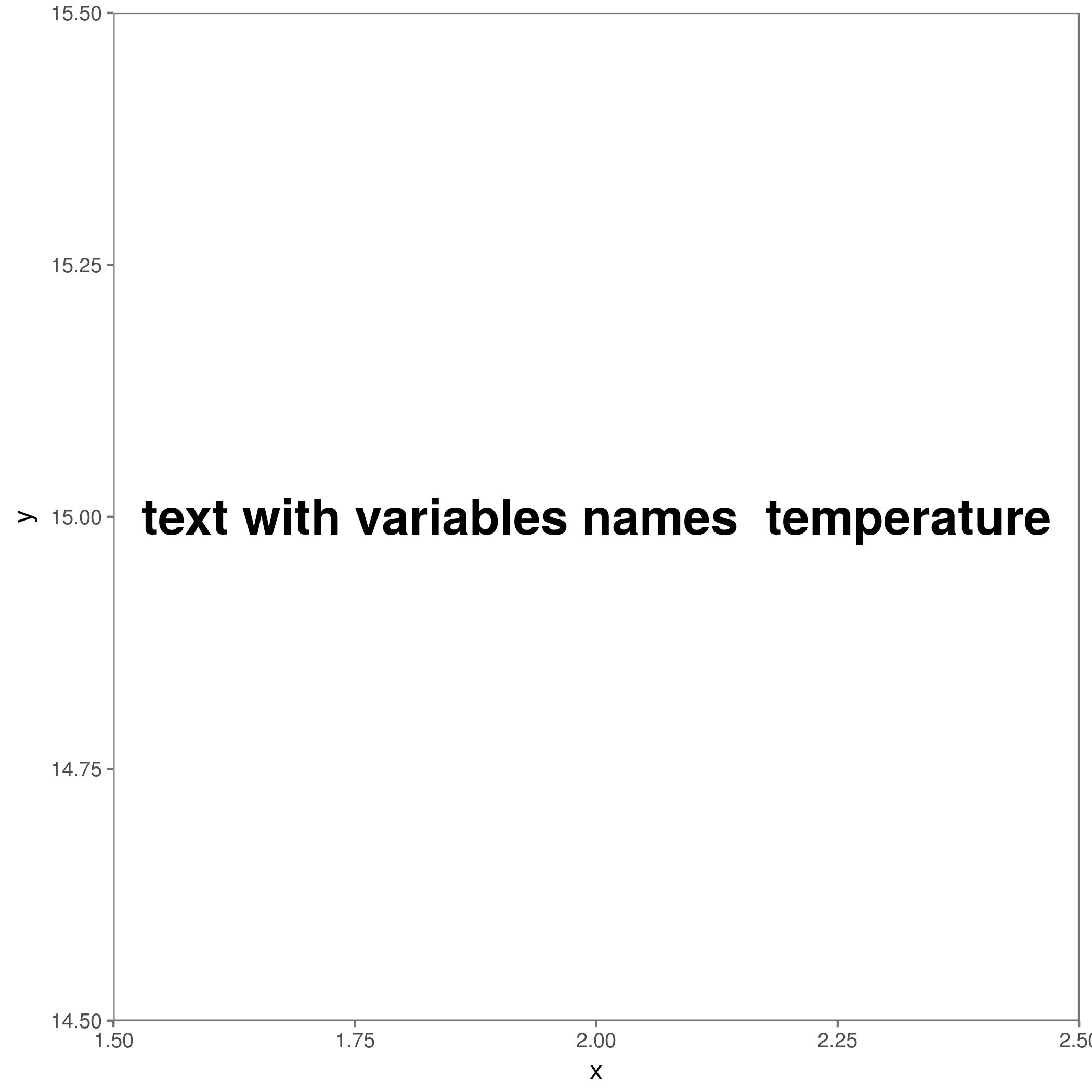
## making a empty graph and save it
ggplot() +
geom_text() +
annotate("text", label = paste0("text with variables names ",i ), x = 2, y = 15, size = 8, fontface = "bold") +
theme_few() +
ggsave(paste0("variables", "_", i,".pdf"))
setwd(mainDir) #here very important to set the mainDir working directory again
}
Here it will check for three different paths in the same subfolder
results RandomForest
/Project X/Analyses/results RandomForest/temperature
/Project X/Analyses/results RandomForest/slope
/Project X/Analyses/results RandomForest/patterns
and save in each folder a file names varialbes_temperature.pdf (or slope.pdf
or patterns.pdf) in the folder temperature (or slope or patterns).
The outputs figures should look like this: